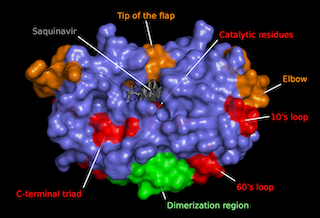
| HIV-1 Protease regions
Molecular rendering of protease dimer surface (slate blue) coupled to the
antiretroviral Saquinavir (grey) highlighting protein domains. Crystallographic
model generated through x-ray diffraction crystallography by Kovalevsky AY, 2010 (PDB:3OXC).
|
 |
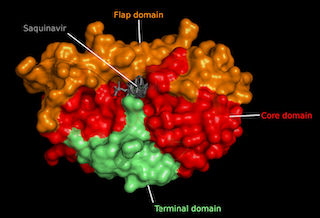
| HIV-1 Protease domains
Molecular rendering of protease dimer surface (slate blue) coupled to the
antiretroviral Saquinavir (grey) highlighting protein domains. Crystallographic
model generated through x-ray diffraction crystallography by Kovalevsky AY, 2010 (PDB:3OXC).
|
 |
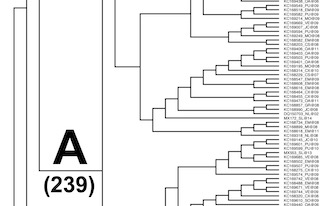
| Phylogeny of Mexican HIV-1 Protease-encoding nucleotide sequences
Phylogeny reconstructed using a Markov Chain Monte Carlo algorithm
based Bayesian analysis suite (BEAUti and BEAST) using a general
time reversible substitution model and strict clock for 777 Mexican
HIV-1 nucleotide sequences. |
 |
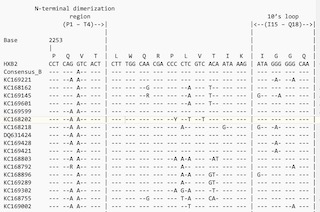
| Mexican HIV-1 Protease-encoding nucleotide sequence alignments
BBEdit text multiple alignment of the 777 Mexican HIV-1 nucleotide sequences along with HXB2
reference and consensus sequences for group M subtypes A1, A2, B, C, D,
F1, F2, G & H and group O as performed using Clustal Omega
and reformatted for unanimity to HXB2 reference using SURe. |
 |
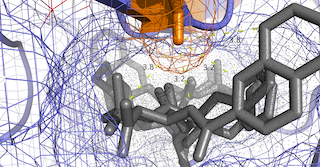
| Protease inhibitor resistance mutations in local HIV-1 Isolates
Primary and secondary protease inhibitor resistance mutations seen in 51
Mexican HIV-1 isolates are mapped unto a crystallographic model
of a HIV-1 protease dimer coupled to the antiretroviral Saquinavir
protease-inhibitor generated through x-ray diffraction crystallography (PDB:3OXC).
|
 |
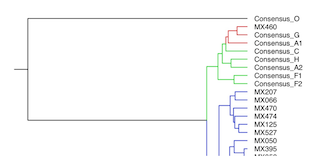
| Phylogeny of local HIV-1 Protease-encoding nucleotide sequences
Phylogeny reconstructed using a Markov Chain Monte Carlo algorithm
based Bayesian analysis suite (BEAUti and BEAST) using a general
time reversible substitution model and log normal relaxed clock for 51 Mexican
HIV-1 nucleotide sequences plus group M (subtypes A1, A2, B, C, D, F1, F2, G and H)
and group O (as an outlier) consensus sequences. |
 |
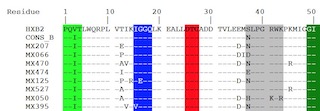
| Local HIV-1 Protease amino acid sequence alignments
Multiple alignment of the 51 Mexican HIV-1 amino acid sequences along with HXB2
reference and consensus sequences for group M subtypes A1, A2, B, C, D,
F1, F2, G & H and group O as performed using Clustal Omega
and reformatted for unanimity to HXB2 reference using SURe. |
 |
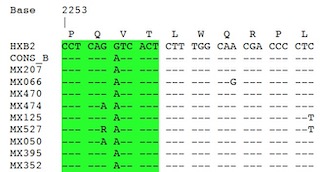
| Local HIV-1 Protease-encoding nucleotide sequence alignments
Multiple alignment of the 51 Mexican HIV-1 nucleotide sequences along with HXB2
reference and consensus sequences for group M subtypes A1, A2, B, C, D,
F1, F2, G & H and group O as performed using Clustal Omega
and reformatted for unanimity to HXB2 reference using SURe. |
 |
| Local HIV-1 Vif amino acid sequence alignments
Multiple alignment of the 86 nucleotide sequences was performed using Clustal Omega
and reformatted for unanimity to the HXB2 reference using SURe. |
 |
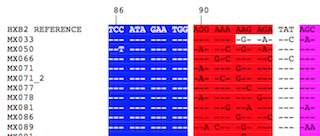
| Local HIV-1 Vif-encoding nucleotide sequence alignments
Multiple alignment of the 86 nucleotide sequences was performed using Clustal Omega
and reformatted for unanimity to the HXB2 reference using SURe. |
 |
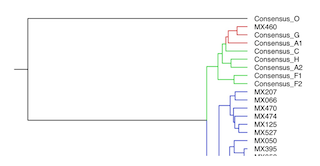
| Phylogeny of Mexican HIV-1 Vif-encoding nucleotide sequences
Phylogeny reconstructed using a Markov Chain Monte Carlo algorithm
based Bayesian analysis suite (BEAUti and BEAST) using a general
time reversible substitution model and strict clock for 77 Mexican
HIV-1 nucleotide sequences plus group M (subtypes A, B, C, D, F1, F2, G and H)
and group O (as an outlier) consensus sequences. |
 |
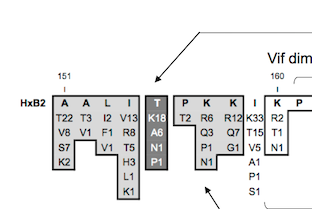
| Mexican Vif protein sequence alignment summary
Protein domains and functional regions are shaded to highlight functional regions
and sites. These include tryptophans (W) involved in APOBEC3G binding, APOBEC3-protein
family binding domains, the nuclear localization inhibitory signal, the MAPK
phosphorylation sites, CBFβ interaction sites, the Cullin5 Zinc-binding domain,
the Elongin B/C (BC) box, protease processing site, known phosphorylation sites,
the Cullin5-box and the dimerization site.. |
 |
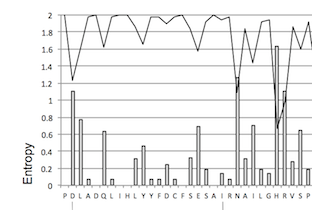
| Mexican Vif protein sequence entropy and amino acid frequency map
The frequency of the most common amino acids observed at each site in the 77 Mexican
HIV-1 Vif sequences is shown in the continuous line while Shannon entropy level at
each site is shown in bar graphs. Boxes indicate protein regions exhibiting low
entropy levels.. |
 |
| HIV pol region sequence analysis and contig assembly
Nucleotide and aminoacid sequences in fasta format and their alignments
for five Human Immunodeficiency Virus type-1 clinical isolates are
included. This page includes links to nucleotide alignments, protein
alignments, fasta files as well as to their original fragment and
contig assembly document. |
 |
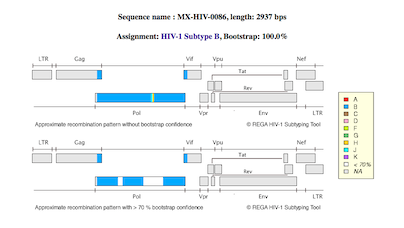
Genomic mapping of contigs unto HXB2 genome pictogram
Topological maps for the five contig clones depicting location of
sequences as generated by REGA HIV-1 Subtyping tool. |
 |
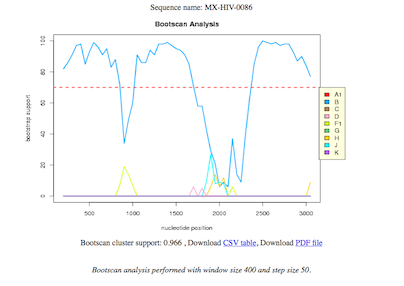
SimPlot analysis and subtyping using bootscan plots of regional homology
Regional homology bootscan plots for the five sequenced contigs.
|
 |
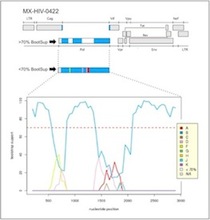
Recombinatorial analysis of cloned and sequenced samples
Recombinatorial analysis incorporating pictogram of HIV genome and
bootscan analysis in a single image. |
 |
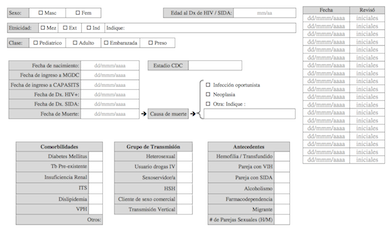
Clinical information capture form (v08) for MGDC-HIV (in spanish)
Includes a 7 page clinical and epidemiological data collection form
in protable document format as used by our lab to summarize individual
medical records. |
 |
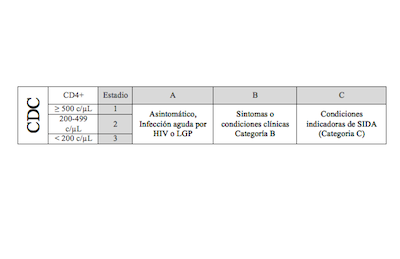
CDC staging table (in spanish)
Includes single page HIV clinical staging table summarizing CDC's
criteria based on both clinical and CD4 cell count data. |
 |